Introduction
Cancer is a heterogeneous disease that affects almost everyone: as a patient, survivor, relative or friend. This page focuses on the key data management challenges, considerations and solutions that are relevant across all stages of the patient’s journey from cancer prevention, diagnosis and treatment to assessment of patient outcomes and monitoring those at follow-up visits.
Each stage of the patient journey has different associated data types, a number of technical, ethical, legal and organisational challenges. In this page we focus on the management of human health data generated from patients diagnosed with both solid and liquid tumors (oncology or hematology). Data might be collected by a number of different means, e.g. from clinical trials and non-interventional studies (NIS) or real-world data (RWD) from observational studies (e.g. registries) or captured in electronic health record (EHR) hospital systems during primary care.
Efficient data management and interoperability are essential for ensuring timely and accurate diagnoses and treatment while maintaining compliance with ethical and regulatory standards. In the following sections we will address data management best practices, considerations and possible solutions for effective management of data in each stage (Figure1) of the individual patient’s journey.
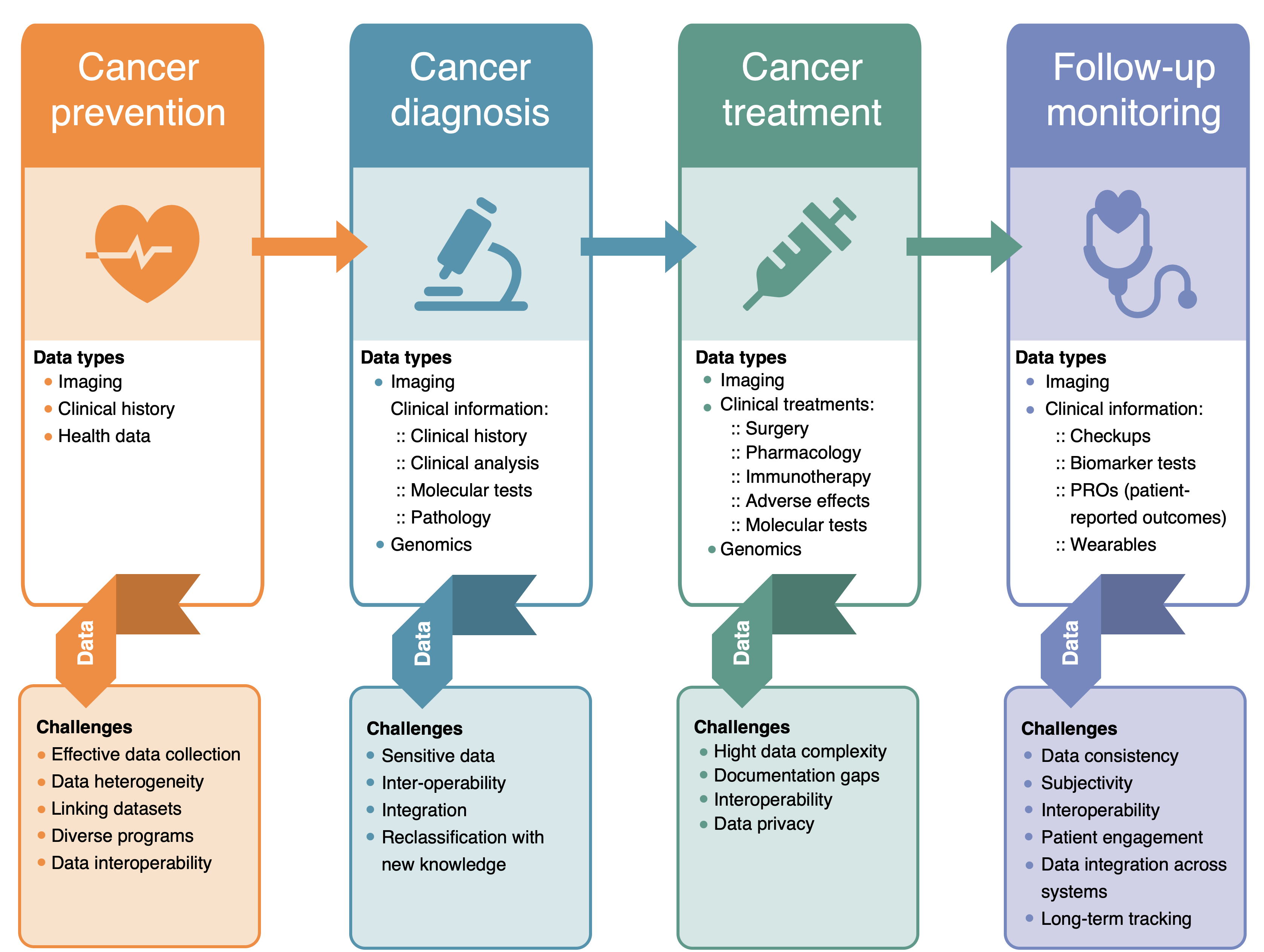
Figure 1. Cancer data management challenges at different stages of patient journey.
Cancer prevention
Description
Primary prevention data
Primary prevention in cancer care refers to strategies aimed at reducing the risk of developing cancer. Data from cancer registries (which includes spatial patterns of cancer incidence, as well as stage, survival and mortality) in combination with genetic predisposition and/or exposome data (including exposure to environmental factors and socio-economic characteristics) can be used to identify risk factors for developing cancer. These cancer registries are information systems designed for the collection, storage, and management of data on persons with cancer and play a critical role in cancer research, surveillance, cancer prevention and control interventions. Key challenges include heterogeneity in data collection and integrating diverse datasets from different sources, e.g. linkage of exposome data to the health data from cancer registries.
Secondary prevention data
Secondary prevention in cancer care focuses on early detection and intervention to identify cancer at an early stage when it is more treatable and potentially curable. Survival rate improvement in most major tumour types depends on early detection, which has prompted screening programs in many European countries. These programs produce highly relevant datasets for further (data-driven) research on early cancer diagnostics. This data typically consists of health and bioimaging data, such as mammograms, colonoscopies, or blood tests. Most of this data contains personal health information and must be managed in compliance with privacy regulations such as GDPR.
Key challenges include integrating diverse datasets and ensuring data accuracy since the screening programs could be organised on national or regional level. Additionally, the risks and benefits of screening programs must be balanced.
Considerations
Primary prevention data
- Consider local cancer registries in the different European countries as they can be organised locally, regionally or nationally.
- Think about the health data access procedures which could be different for each cancer registry.
- Bear in mind the interoperability of variables from the exposome data could be suboptimal due to heterogeneous data collection between different sources.
- Linkage between different data types, e.g. exposome and health data, could be non-trivial. Think about the following:
- Does the geographical grid match?
- Does the timestamp of the data correlate?
- Exposome data is considered non-personal data, but once linked to personal data the linked dataset becomes personal data and privacy has to be ensured in compliance with applicable legislation (e.g. GDPR).
Secondary prevention data
- The data access procedure could be different for different data sources. Be mindful to contact the relevant data creators and managers for the relevant access rights.
- Interoperability of data originating from multiple screening programs is not guaranteed.
- For general health data considerations, see Health data page.
- For general bioimaging data considerations, see BioImaging data page.
Solutions
Primary prevention data
- For data management considerations and solutions of Patient-generated health data and Electronic Health Record (EHR) data see the Health data page.
- Cancer registry data common rules and definitions used within Europe defined by the European Network of Cancer Registries (ENCR).
- Exposome data management recommendations under development by Environmental Exposure Assessment Research Infrastructure (EIRENE-RI).
- Exposome (meta)data definitions used within Europe defined by Eurostat, Euro SDMX Registry.
Secondary prevention data
As there are no commonly accepted data collection standards currently, EOSC4Cancer developed a harmonised codebook for colorectal cancer screening (based on Dutch, Catalan, Italian and Czech screening codebooks), which could be used as a common basis to be extended to other cancer types.
Cancer diagnosis
Description
A cancer patient’s journey starts with a confirmed diagnosis, which involves clinical evaluation, imaging, laboratory tests, and testing of molecular biomarkers by different methods (e.g. immunohistochemistry, next-generation sequencing, in situ hybridisation) in biopsies (e.g. tissue or liquid biopsies) to confirm malignancy and assess tumour characteristics.
Cancer diagnosis is a multi-step process that begins with a patient’s clinical presentation, such as symptoms or incidental findings during routine check-ups or specialised screening programs (e.g. mammography for breast cancer, FIT tests and consequent colonoscopy for colorectal cancer, Pap smears and HPV PCR for cervical cancer). If cancer is suspected, the diagnostic journey typically involves a combination of medical imaging (CT, MRI, PET scans, ultrasound), laboratory tests, and biopsy procedures to confirm malignancy, each step producing a different data type. Integrating diverse data sources, including clinical history, imaging, pathology, and genomic data, allows for a more comprehensive understanding of the disease and personalised treatment strategies.
Cancer diagnosis relies on data from multiple sources that are also often used at other stages of the cancer patient’s journey (prevention, treatment, follow-up):
- Imaging (MRI, CT, PET scans) provides tumor size, location, and spread
- Pathology (biopsy analysis) confirms tumor type and molecular characteristics
- Genetic/Genomic profiling can identify tumor genomic alterations relevant for molecularly matched therapies, pharmacogenomic biomarkers relevant for drug metabolism, germline alterations
- Clinical data (patient history, symptoms, lab tests) provide context on overall health and treatment history
Managing cancer data for diagnosing and determining the best treatment for localized tumors presents several challenges, as it requires working with a wide range of sensitive patient data, coming from different departments/sources, including clinical records, radiological images (radiology), histopathological evaluation (pathology) and genomic profiles (pathology or other specialised laboratory). This makes interoperability and data integration essential to enable a holistic approach to cancer care. Consequently, data management must be precise to ensure that healthcare professionals have accurate and comprehensive information for tumour identification and treatment decisions. Data security and compliance with ethical guidelines (such as GDPR) are critical to protecting patient privacy when dealing with personal health records and sensitive tumour data. Furthermore, the need for data to be accessible across different healthcare providers and research institutions adds complexity to the management process.
Considerations
- Are all clinically relevant variables collected using standard vocabularies across data domains including socio-demographics, risk factors, and tumor-specific metadata (e.g. Tumour- Nodes-Metastases (TNM) stage, histology, genomic alterations)?
- Are diagnostic data and images stored in standardised formats (e.g. Digital Imaging and Communications in Medicine (DICOM) for imaging, Variant Call Format (VCF) for genomics) to allow long-term usability and reanalysis?
- Is there a data management system in place to ensure interoperability between different data types (e.g. imaging, molecular, and health records)?
- Are there AI-based tools or decision-support systems integrated into the workflow to assist oncologists in making diagnostic decisions?
Solutions
- Utilize structured clinical data models and interoperability frameworks (e.g. HL7 FHIR, OMOP-CDM, Digital Imaging and Communications in Medicine (DICOM) and XNAT for imaging) to facilitate the integration of multi-modal diagnostic data such as clinical, imaging, and molecular data.
- Implement version-controlled patient records using EHR systems that support updates based on evolving classification standards and cancer-specific coding dictionaries for diagnosis (e.g. ICD-O-3 Coding Materials or Systematized Nomenclature of Medicine Clinical Terms (SNOMED CT) for cancer diagnosis and topography, Tumour, Nodes, Metastases (TNM) cancer staging system staging system, WHO Classification of Tumours).
- Utilize secure repositories and specialised cancer clinical data management systems (e.g. REDCap, Informatics for Integrating Biology & the Bedside (i2b2), cBioPortal) that comply with GDPR, HIPAA, and other regulatory frameworks.
- Implement standardised consent forms and patient data governance frameworks, such as GA4GH’s Data Use Ontology (DUO), to allow data sharing while maintaining privacy.
- Store raw sequencing and imaging data in cloud-based or institutional repositories (e.g. The European Genome-phenome Archive (EGA), dbGAP, Sequence Read Archive, The Cancer Imaging Archive (TCIA) for imaging) to allow reanalysis when new prognostic markers emerge.
- Adopt federated learning approaches (e.g. Federated EGA, Federated EHR Learning Models) to enable collaborative research without transferring sensitive patient data.
- Integrate AI-based imaging tools (e.g. PathAI, Qure.ai, Paige AI) for radiology and pathology analysis to assist in detecting subtle cancer features and ensure adherence to standards (e.g. DOME-ML, Transparent Reporting of a Multivariable Prediction Model), avoiding biases in cancer diagnosis.
Cancer treatment
Description
Cancer treatment varies depending on the type and stage of cancer (e.g. locally advanced, metastatic disease), as well as the overall health and preferences of the patient. The use of advanced diagnostic techniques such as PET-CT/MRI, molecular profiling (e.g. next-generation sequencing, comprehensive genomic profiling (CGP), whole genome sequencing (WGS) and liquid biopsies (e.g. ctDNA) has tremendously increased the data density and complexity to be dealt with at this stage of disease.
Cancer treatment employs a wide range of data types, such as patients’ therapeutic regimens, including surgery techniques, stem cell transplantation, radiotherapy, systemic therapies (e.g. hormone, chemotherapy, immunotherapy and targeted therapies) as well as imaging data, biomarker assessments, responses to therapies data, clinical trial outcomes, drug efficacy, and adverse reactions. Cancer treatment data is commonly associated with further clinical data and patients’ information. Due to their sensitive nature, the data must be managed following ethical guidelines, data protection laws, and FAIR (Findable, Accessible, Interoperable, and Reusable) principles.
Although cancer treatment data is crucial for developing personalised medicine approaches, improving patient outcomes and advancing research, comprehensive documentation of cancer treatment data remains limited in cancer registries and public datasets. This challenge is often due to data privacy regulations, ethical concerns, and varying reporting standards, which highlight disparities arising from resource limitations, national database structures, and language barriers. In addition, while cancer treatment data publication has increased, it remains inconsistent due to the lack of data standardisation along with sparse ontologies. The increasing use of electronic health records across western countries, along with standardised cancer classification systems (e.g. WHO, ICD, CAP), staging systems (e.g. Tumour, Nodes, Metastases (TNM) cancer staging system ), and pioneering drug (e.g. DRON PDRO) and side effects (e.g. OAE) ontologies, facilitates data collection. However, clear guidelines for cancer treatment data collection and tools for unified analysis still need to be developed.
Considerations
- Do you use human data? You can find more information on the Human data page.
- Are the required clinical variables related to the treatment available?
- How will clinical variables be integrated with molecular or imaging data?
- Which resources are available for downloading and analysing cancer treatment data?
- Where can you access standard-of-care cancer clinical guidelines?
- How to access cancer treatment data from clinical trials or side effect registries?
- How to propose cancer treatments based on cancer multi-omics data?
Solutions
In order to obtain information about oncological clinical practice guidelines several medical societies provide guidance:
- European Society of Medical Oncology (ESMO)
- American Society of Clinical Oncology (ASCO)
- National Comprehensive Cancer Network (NCCN)
A more unified approach to cancer treatment data collection is crucial for improving outcome analysis and supporting all stakeholders. To support this aim, several consortia and institutions provide annotated reference datasets with cancer treatment data:
Reference databases and platforms:
- The European Genome-phenome Archive (EGA) : Service for permanent archiving and sharing of personally identifiable genetic, phenotypic, and clinical data.
- cBioPortal: Multidimensional genomics data with treatment annotations.
- ICGC-ARGO: Comprehensive clinical and genomic data for >100,000 patients.
- AACR-GENIE: Cancer genomic dataset.
- MSK-CHORD: Molecular and clinical data for cancer treatment analysis.
- Hartwig Medical Foundation (HMF): Multi-omic data for >7000 patients.
- WAYFIND-R: Real-world clinical/genomic data from patients diagnosed with solid tumours across geographies.
- European Cancer Imaging Initiative (EUCAIM): Federated platform for Cancer image data.
- All of Us: US-based patient health data.
- UK Biobank: UK-based longitudinal study of 500,000 UK individuals; multi-level data across a range of diseases.
- The Cancer Imaging Archive (TCIA): Imaging data with metadata.
- Genomics of Drug Sensitivity in Cancer (GDSC)): Drug sensitivity data from 1000 cancer cell lines.
- Cancer Therapeutics Response Portal (CTRP): Genetic and drug sensitivity relationships.
Drug and trial public repositories:
- National Cancer Institute (NCI) treatment drugs
- European Medicines Agency’s (EMA) medicine finder
- ClinicalTrials.gov
- DrugBank
- Drug-Gene Interaction Database (DGIdb)
- DrugMap
- Therapeutic Target Database (TTD)
- Cancer Pharmacogenomics
- OncoKB
- SIDER
Genomics & multi-omics resources:
- MTB Portal: provides a general framework to interpret the functional and predictive relevance of a given list of gene variants in interactive reports.
- PanDrugs: a platform to prioritize cancer drug treatments according to individual multi-omics data (SNVs, CNVs and gene expression).
- Cancer Genome Interpreter: flags genomic biomarkers of drug response with clinical relevance.
- Clinical Interpretations of Variants in Cancer (CIViC): a free resource to identify the best cancer treatment options based on DNA alterations.
- Topograph: Therapy-Oriented Precision Oncology Guidelines for Recommending Anti-cancer Pharmaceuticals.
Monitoring of outcomes during follow-up visits
Description
The follow-up phase in cancer care is a critical component of comprehensive patient management, ensuring long-term monitoring and well-being of cancer survivors. This stage focuses on assessing treatment outcomes, detecting potential recurrences, managing long-term side effects, and enhancing the overall quality of life. Effective follow-up strategies integrate not only systematic clinical evaluations, which include routine medical visits, imaging exams (e.g. MRI, CT, PET), and biomarker testing (e.g. CEA, PSA, ctDNA), but also patient-reported outcomes (PROs).
In this context, the increasing adoption of digital health technologies, including wearable devices and mobile health applications, as well as Artificial Intelligence and predictive analytics, has transformed post-treatment monitoring. On one hand, it has enabled real-time remote tracking of health metrics (e.g. physical activity, heart rate, sleep patterns etc), facilitating early detection of potential complications and on the other hand help anticipate complications and tailor follow-up schedules to individual patients’ needs. Both scenarios lead to generation of diverse data types. Additionally, cancer registries (CRs) and clinical trial databases play a fundamental role in storing longitudinal data on disease progression, survival rates, and treatment efficacy, allowing researchers to analyse trends, identify recurrence risk factors, and refine personalised follow-up guidelines.
However, due to the wide heterogeneity of data types, sources, and healthcare systems achieving seamless interoperability and standardisation of follow-up data, to support individualised patient management and optimise data reuse in cancer research remains a major challenge. In addition, data collection and management at this stage presents other challenges, including: (i) the sensitive nature of the data, requiring strict adherence to regulatory and ethical frameworks, (ii) the lack of consistency and/or quality of patient follow-up information, and (iii) the lack of standardisation and inherent subjectivity in survivorship quality-of-life data, influenced by patient perception, reporting methods, and assessment tools.
Considerations
Different considerations should be taken into account depending on the type of data being managed:
- Use specific standards and methods to extract and transform data included in the Electronic Health Record (clinical data, diagnoses, demographics, procedures, medications, vital signs, laboratory results). For Considerations towards improved reuse of EHR refer to the section in the Health data page.
- Considerations for managing imaging data (and histopathological data), binary files,as well as the associated metadata can be found in the Bioimaging data page.
- For human genomic data, established research ethical guidelines and legislations must be followed as described in the Human data page.
- Since health data falls under the “special category of data” as defined by the GDPR, strict guidelines and considerations must be followed when handling this information covered in the GDPR compliance and Data Sensitivity pages of the RDMkit.
-
For PROs, to collect data directly from cancer patients and/or survivors, follow the considerations listed on the health data page. Additionally, since these PROs focus on quality-of-life and are inherently subjective, additional considerations must be addressed:
- Are questionnaires designed to minimise ambiguity and ensure that all patients interpret questions in a similar way?
- Are there methods in place to differentiate between true changes in quality of life and variations due to individual perception or recall bias?
- Have statistical or methodological approaches been considered to adjust for subjectivity in self-reported data?
- How is the potential discrepancy between patient-reported outcomes and clinician assessments addressed?
-
For data collected from wearable devices and mobile applications, the follow considerations should be taken into account:
- Are the wearable devices and mobile applications validated to provide accurate real-time measurements?
- How is the data quality ensured, considering potential sensor calibration issues, environmental factors, or user error?
- Are there mechanisms in place to handle missing or incomplete data, such as when the device is not worn or battery levels are low?
- How are transient fluctuations in health metrics differentiated from clinically significant changes?
- Are patient-specific factors incorporated into the analysis to improve data interpretation?
- Is there a system for aggregating data from multiple devices or platforms to create a comprehensive view of the patient’s health metrics over time?
-
To address the potential lack of consistency and/or quality in patient follow-up information, particularly over the long term, the following considerations should be taken into account:
- How will data consistency be maintained if patients change healthcare providers, devices, or platforms over time?
- Are there standardised processes in place to ensure that follow-up data from different sources can be seamlessly integrated and compared?
- Is there a plan to handle potential gaps in data, such as missed follow-up appointments or missing reports?
- What strategies are in place to encourage continuous patient engagement and adherence to follow-up protocols?
- Are there periodic checks or audits in place to validate data quality and identify potential discrepancies or inconsistencies?
Solutions
- To address the challenges associated with the monitoring of outcomes during follow-up visits, the following solutions are proposed:
- Adopt common data models and standards to ensure consistency and interoperability of clinical data across institutions.
- Regarding data included in the EHR, for standards that are likely to be present as part of such systems, common data models that can facilitate data sharing, and tools for data integration, refer to the section in the Health data page.
- Regarding imaging data, for standard (meta)data formats, management platforms, ontologies resources, and image repositories, refer to the Bioimaging page.
- Use cancer-specific standard terminologies, ontologies, and reference databases for coding diagnosis related data (e.g. ICD-O-3 Coding Materials, WHO Classification of Tumours, Tumour, Nodes, Metastases (TNM) cancer staging system), treatment related data (e.g. Cancer Therapeutics Response Portal (CTRP), Clinical Interpretations of Variants in Cancer (CIViC), Therapeutic Target Database (TTD)) and follow-up related data (e.g. Medical Dictionary for Regulatory Activities (MedDRA), Eastern Cooperative Oncology Group (ECOG), PET Response Criteria in Solid Tumors (PERCIST)).
- Utilise GDPR compliant EDC systems that support capture of PROs (refer to the Health data page and structured formats (e.g. FHIR QuestionnaireResponse).
- Employ validated and standardised instruments for quality-of-life assessment (e.g. European Organisation for Research and Treatment of Cancer (EORTC) Quality of Life Questionnaires (QLQ), Patient-Reported Outcomes Measurement Information System (PROMIS)) to reduce variability in interpretation and improve comparability.
- Use certified, clinically validated devices and apps to ensure data reliability and regulatory compliance (e.g. CE-marked, FDA-approved tools). Employ data aggregation platforms (e.g. Apple HealthKit, Google Fit, Open mHealth) that support cross-device integration and longitudinal monitoring.
- Define clear data governance policies for longitudinal data capture and ensure data traceability.
- Establish patient engagement protocols to support consistent reporting and minimise data loss over time. Define standardised follow-up templates to optimise data completeness.
Related pages
More information
Links to FAIRsharing

FAIRsharing is a curated, informative and educational resource on data and metadata standards, inter-related to databases and data policies.
Training
Tools and resources on this page
| Tool or resource | Description | Related pages | Registry |
|---|---|---|---|
| AACR-GENIE | Real-world cancer genomic dataset aggregated from multiple institutions. | ||
| All of Us | NIH research program with diverse patient health, genetic, and lifestyle data. | Training | |
| Apple HealthKit | A framework for health and fitness data integration on Apple devices. | ||
| Cancer Genome Interpreter | Cancer Genome Interpreter (CGI) is designed to support the identification of tumor alterations that drive the disease and detect those that may be therapeutically actionable. | Human data | Tool info |
| Cancer Pharmacogenomics | Pharmacogenomics knowledge base including cancer pharmacogenomic information. | ||
| Cancer Therapeutics Response Portal (CTRP) | Resource to explore relationships between cancer cell line genotypes and drug response. | ||
| cBioPortal | A platform for exploring, analysing, and visualising cancer genomics data. | Tool info Standards/Databases Training | |
| Clinical Interpretations of Variants in Cancer (CIViC) | A community knowledgebase for expert crowdsourcing the clinical interpretation of variants in cancer | Tool info | |
| ClinicalTrials.gov | ClinicalTrials.gov is a resource depending on the National Library of medicine which makes available private and public-funded clinical trials. | Toxicology data | Standards/Databases |
| Data Use Ontology (DUO) | DUO allows to semantically tag datasets with restriction about their usage. | Human data Ethical aspects | Standards/Databases Training |
| dbGAP | The database of Genotypes and Phenotypes (dbGaP) archives and distributes data from studies investigating the interaction of genotype and phenotype in Humans | Human data | Tool info Standards/Databases Training |
| Digital Imaging and Communications in Medicine (DICOM) | A standard for medical imaging data storage and transmission. | Standards/Databases | |
| DOME-ML | Data, Optimisation, Model and Evaluation in Machine Learning (DOME-ML) is a set of community guidelines, recommendations and checklists for supervised ML validation in biology. | Machine learning | Standards/Databases |
| Drug-Gene Interaction Database (DGIdb) | Drug-Gene Interaction Database linking drugs to their target genes. | Tool info | |
| DrugBank | Comprehensive database containing information on drugs and drug targets. | Tool info Standards/Databases Training | |
| DrugMap | Database for drug mechanisms, targets, pathways, and indications. | Tool info Standards/Databases | |
| Eastern Cooperative Oncology Group (ECOG) | A scale for assessing patient functional status. | ||
| European Cancer Imaging Initiative (EUCAIM) | A federated platform for cancer imaging data. | ||
| European Medicines Agency's (EMA) medicine finder | European Medicines Agency's (EMA) portal for authorised medicines in the EU. | ||
| European Organisation for Research and Treatment of Cancer (EORTC) Quality of Life Questionnaires (QLQ) | A set of questionnaires developed to assess the quality of life of cancer patients. | ||
| FDA-approved tools | Various digital health tools approved by the FDA for medical use (e.g., wearables, software, diagnostics). | ||
| Federated EGA | The Federated EGA is an infrastructure built upon the European Genome-phenome Archive (EGA), an EMBL-EBI and CRG data resource for secure archiving and sharing of human sensitive biomolecular and phenotypic data resulting from biomedical research projects. | Training | |
| Genomics of Drug Sensitivity in Cancer (GDSC) | Database of drug sensitivity and genomic data from cancer cell lines. | ||
| Google Fit | A health-tracking platform by Google for collecting and accessing fitness data. | ||
| Hartwig Medical Foundation (HMF) | Multi-omic cancer dataset of >7000 patients, including whole genome sequencing and clinical outcomes. | ||
| HL7 FHIR | A standard used for health care data exchange. | Health data | |
| ICD-O-3 Coding Materials | A classification system for oncology, used in cancer registries and pathology reporting. | ||
| ICGC-ARGO | Integrated clinical and genomic data for over 100,000 cancer patients from the International Cancer Genome Consortium. | ||
| Informatics for Integrating Biology & the Bedside (i2b2) | An open-source data warehouse for clinical and translational research. | Training | |
| Medical Dictionary for Regulatory Activities (MedDRA) | A single standardised international medical terminology. | Standards/Databases | |
| MSK-CHORD | Platform from Memorial Sloan Kettering providing molecular and clinical data to support precision oncology. | ||
| MTB Portal | Provides a general framework to interpret the functional and predictive relevance of a list of gene variants through interactive reports. | ||
| National Cancer Institute (NCI) treatment drugs | A database of cancer-related drugs, including descriptions, uses, and approvals. | Training | |
| OMOP-CDM | OMOP is a common data model for the harmonisation for of observational health data. | TransMed Data quality | |
| OncoKB | Precision oncology knowledge base that annotates the effects and treatment implications of somatic mutations in cancer. | ||
| Open mHealth | An open-source Clinical Interpretations platform for integrating mobile health data to improve healthcare insights. | Standards/Databases | |
| Paige AI | AI solutions for digital pathology in cancer detection. | ||
| PanDrugs | Platform to prioritise cancer drug treatments based on individual multi-omics data including SNVs, CNVs, and gene expression. | Tool info Standards/Databases Training | |
| PathAI | AI-powered pathology tools for cancer diagnosis and research. | ||
| Patient-Reported Outcomes Measurement Information System (PROMIS) | A set of person-centred measures that evaluates and monitors physical, mental, and social health in adults and children. | ||
| PET Response Criteria in Solid Tumors (PERCIST) | A standardised method for assessing tumor response via PET imaging. | ||
| Qure.ai | AI-based medical imaging analysis for radiology and oncology. | Tool info | |
| REDCap | REDCap is a secure web application for building and managing online surveys and databases. While REDCap can be used to collect virtually any type of data in any environment, it is specifically geared to support online and offline data capture for research studies and operations. | TransMed Health data Data quality Identifiers | Tool info Training |
| Sequence Read Archive | Sequence Read Archive (SRA) data, available through multiple cloud providers and NCBI servers, is the largest publicly available repository of high throughput sequencing data. | Epitranscriptome data Single-cell sequencing | Standards/Databases Training |
| SIDER | Side Effect Resource containing information on marketed medicines and their recorded adverse drug reactions. | Tool info | |
| Systematized Nomenclature of Medicine Clinical Terms (SNOMED CT) | Terminology standard consisting of codes, terms, synonyms, and definitions used in clinical documentation and reporting. | Health data | Standards/Databases |
| The Cancer Imaging Archive (TCIA) | A repository of medical images related to cancer. | Standards/Databases | |
| The European Genome-phenome Archive (EGA) | EGA is a service for permanent archiving and sharing of all types of personally identifiable genetic and phenotypic data resulting from biomedical research projects
|
CSC TSD Human data Virology Data interlinking Data publication | Tool info Standards/Databases Training |
| Therapeutic Target Database (TTD) | Database of known and explored therapeutic targets and their related drugs. | Tool info Standards/Databases | |
| Transparent Reporting of a Multivariable Prediction Model | Guidelines for reporting predictive model studies in medicine. | Standards/Databases | |
| Tumour, Nodes, Metastases (TNM) cancer staging system | A cancer staging system that describes the size of the tumor (T), lymph node involvement (N), and metastasis (M). | ||
| UK Biobank | Large-scale biomedical database with genetic, clinical, and lifestyle data from UK participants. | Tool info Training | |
| Variant Call Format (VCF) | A common file format that contains information about variants found at specific positions in a reference genome. | Plant sciences | Standards/Databases |
| WAYFIND-R | A federated real-world evidence platform containing clinical and genomic data from patients with solid tumours. | ||
| WHO Classification of Tumours | The standard for diagnostic oncology, defining tumor types and classifications. | ||
| XNAT | Open source imaging informatics platform. It facilitates common management, productivity, and quality assurance tasks for imaging and associated data. | TransMed XNAT-PIC Bioimaging data | Tool info |
